Engineering Custom Arrays for Cancer and Autoimmunity Diagnostics
(Neuro panel, overlapping peptide panel, PTM modified autoantigen panel)
Lead: Yewei Ma
Team Members: Sofia I Cantu, Valeria V Espinosa
Collaborator: Dr. Weiyi Peng, Dr. Tianfu Wu, Dr. Prithvi Raj
Project Summary:
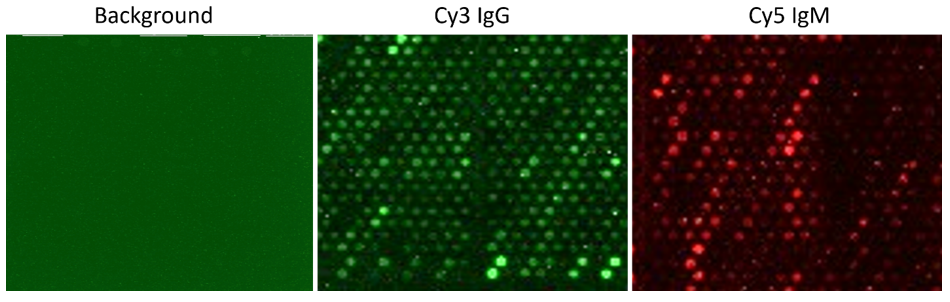
Traditional serological tests such as ELISAs and immunoblots examine only a limited number of antigens, offering a restricted view of the immune response. Custom-engineered peptide arrays provide a multiplexed alternative, enabling simultaneous interrogation of thousands of epitopes. These platforms capture detailed autoantibody repertoires and support biomarker discovery, patient stratification, and therapy monitoring, particularly in cancer and autoimmune diseases.
Newer custom arrays are being engineered to enable identification of linear and cross-reactive epitopes, neoantigen recognition, therapy-induced epitope spreading, and PTM-specific autoantibodies linked to disease subtypes and immune tolerance breakdown.
What is already known in the field?
Peptide and protein arrays have already supported discovery of conformational vs linear epitopes. PTMs create neo-epitopes recognized in many diseases, as exemplified by anti-citrullinated peptide antibodies in Rheumatoid Arthritis, anti-phosphorylated epitopes in neurodegeneration and cancer, and others.
What is new?
Current arrays do not allow sufficient interrogation of PTMs and peptide epitopes that may be relevant across different diseases. Advances in array technology now enable micro- to nano-scale features with higher signal-to-noise, controlled PTM synthesis, and high-density arrays containing up to tens of thousands of peptides.
Why is this important?
Custom array technologies improve diagnostic precision and coverage by enabling comprehensive autoantibody profiling beyond single-antigen assays, increasing sensitivity for early disease detection and improving discrimination between differential diagnoses. They may also uncover novel, patient-specific epitopes – including PTM-derived neo-epitopes – supporting personalized disease subtyping, monitoring and even tailored therapies.
Ongoing/future steps:
Translating research arrays into commercial diagnostic tools, establishing normative databases, and validating cutoff thresholds and reproducibility. Using machine learning to predict immunodominant epitopes, customize antigen panels automatically and identify epitopes predictive of disease severity, progression or treatment response.