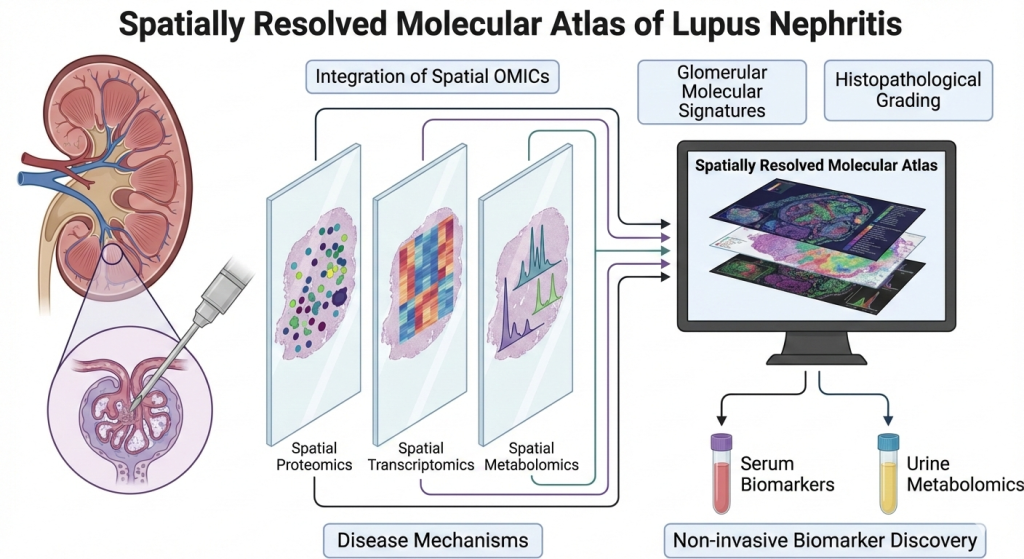
Spatial Proteomic, Transcriptomic, and Metabolomic Investigations of Lupus Nephritis
Lead: Crosslee Titus
Team Members: Wasim Akram, Akanksha Reddy, Mohammad Dauok, Munim Ahmed, Aalekhya Biswas, Ayesha Bhudwani
Collaborators: Dr. Ramesh Saxena, Dr. Luan Truong, Dr. Rohith Appalaneni, Dr. Qi Cai
Project Summary:
Lupus nephritis (LN) is a renal manifestation of Systemic Lupus Erythematosus, an autoimmune kidney disease in which molecular changes within glomeruli and tubulointerstitial compartments drive disease activity, progression, and irreversible fibrosis. In this project, we integrate spatial proteomics, spatial transcriptomics, and spatial metabolomics on adjacent tissue sections from LN kidney biopsies to generate a spatially resolved molecular atlas of LN. By analyzing and co-registering the signatures from multiple OMICs platforms within the same anatomical regions, we aim to identify disease-relevant pathways, cell-specific responses, and non-invasive biomarkers that reflect intrarenal pathology.
What is already known in the field?
Prior studies using bulk transcriptomics, proteomics, or single-cell RNA sequencing have identified immune activation, interferon signaling and fibrotic pathways in LN. However, these approaches lack spatial context and cannot link molecular changes to specific glomerular states, cellular neighborhoods, or histopathologic features such as activity and chronicity indices. Similarly, metabolomic studies of LN urine and serum identify dysregulated metabolites but cannot trace back their renal cellular sources.
What is new?
We have conducted spatial OMICs (transcriptomics, proteomics and metabolomics) on adjacent sections from kidney biopsies of varying disease severities, enabling glomerulus-level integration of gene / protein expression and metabolite profiles with histopathological grading. Glomerular molecular signatures are correlated with LN activity and chronicity indices and cross-referenced with non-invasive urine metabolomics and serum proteomics to identify non-invasive biomarkers reflective of intrarenal molecular activity.
Why is this important?
This work directly addresses a major gap in LN research, the lack of spatial molecular biomarkers that reflect disease mechanisms rather than downstream injury. By linking spatial molecular changes in glomeruli to urinary/serum biomarkers, this project establishes a mechanistic framework for non-invasive disease monitoring and enables patient stratification based on underlying biology rather than histology alone.
Ongoing/Future Steps
- Expand the panel size of spatial proteomics.
- Co-registering ROIs across OMICs platforms
- Validate candidate biomarkers in independent serum/urine cohorts
- Link spatial molecular signatures to treatment response and longitudinal outcomes